An algorithm that can speed up by years the ability to identify from among thousands of possibilities, two or more drugs that work synergistically against a problem like cancer or a viral infection has been developed by bioinformatics experts.
The new algorithm enables investigators to use large existing databases with information about how one cancer drug changed the gene expression of a particular breast cancer cell line, and how well it killed the cell, then mathematically combine those results with the impact of another drug to see if they could work better together, says Dr. Richard McIndoe, director of the MCG Center for Biotechnology and Genomic Medicine.
While the algorithm does not immediately make available the kind of information that would set a clinical trial in motion, it does speed up the path to the trials, he says.
Drug combination therapies can improve drug efficiency, reduce drug dosage (and related toxicity) and overcome drug resistance in cancer treatments,” the investigators write in the journal PLOS ONE, and is becoming an important tool in cancer treatment. But given the number of drugs and drug combinations available, there are not efficient, effective ways to identify the best combinations.
And, not all combinations are beneficial, in fact one drug can actually work as an antagonist against the other, effectively blocking or at least reducing its therapeutic impact. The right combination will, by contrast, enhance the impact of the treatment, McIndoe says, which means together they are better at killing cancer cells.
The algorithm also enables synergy between scientists by enabling easily sharing findings which enables even more drugs and cell lines to be evaluated and the database of effective combinations against specific cancers to grow more rapidly.
There are huge existing databases of cell lines which have been treated with one drug to look at the impact on gene expression, before and after treatment, including the Library of Integrated Network-based Cellular Signatures project, to help streamline the large-scale studies like the MCG investigators wanted to do.
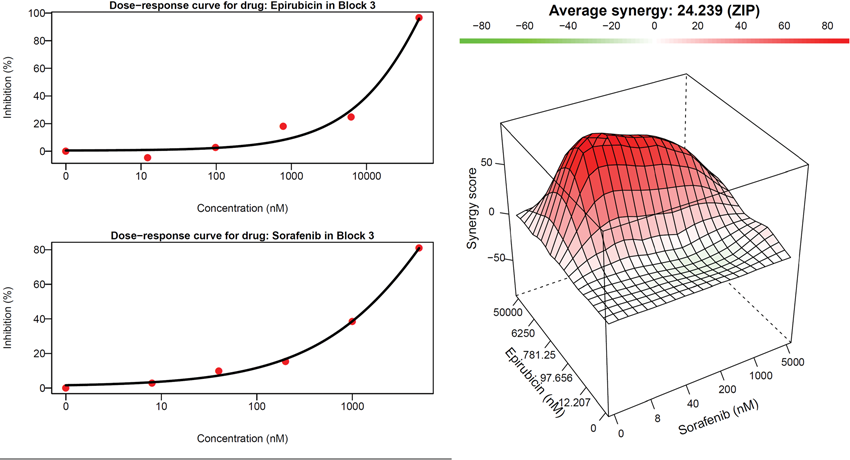
They focused on 57 randomly selected chemotherapy drugs used in the database, looking in detail at the molecular changes each drug produced and tying that to growth rate, meaning how much cancer cell killing the drug produced, then devised a mathematical representation of the molecular changes and the amount of killing for each.
All told there were 1,596 combinations of the 57 cancer drugs they studied. Their algorithm picked 30 top drug combinations and eight were confirmed using a standard statistical model called ZIP, a result far better than chance and far less costly and time consuming than testing the large number of potential drug combinations.










